Researchers develop automated melanoma detector for skin cancer screening
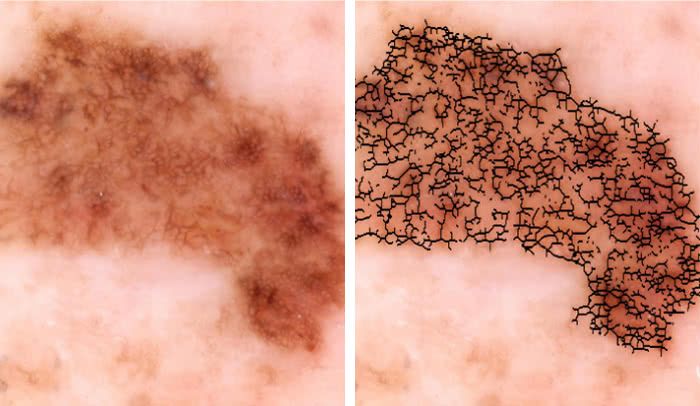
Malignant or benign?: An image of a skin lesion is processed by a new technology to extract quantitative data, such as irregularities in the shape of pigmented skin, which could help doctors determine if the growth is cancerous.
Even experts can be fooled by melanoma. People with this type of skin cancer often have mole-looking growths on their skin that tend to be irregular in shape and color, and can be hard to tell apart from benign ones, making the disease difficult to diagnose.
Now, researchers at The Rockefeller University have developed an automated technology that combines imaging with digital analysis and machine learning to help physicians detect melanoma at its early stages.
“There is a real need for standardization across the field of dermatology in how melanomas are evaluated,” says James Krueger, D. Martin Carter Professor in Clinical Investigation and head of the Laboratory of Investigative Dermatology(opens in new window). “Detection through screening saves lives but is very challenging visually, and even when a suspicious lesion is extracted and biopsied, it is confirmed to be melanoma in only about 10 percent of cases.”
In the new approach, images of lesions are processed by a series of computer programs that extract information about the number of colors present in a growth, and other quantitative data. The analysis generates an overall risk score, called a Q-score, which indicates the likelihood that the growth is cancerous.
Published in Experimental Dermatology, a recent study evaluating the tool’s usefulness indicates that the Q-score yields 98 percent sensitivity, meaning it is very likely to correctly identify early melanomas on the skin. The ability of the test to correctly diagnose normal moles was 36 percent, approaching the levels achieved by expert dermatologists performing visual examinations of suspect moles under the microscope.
“The success of the Q-score in predicting melanoma is a marked improvement over competing technologies,” says Daniel Gareau, first author of the report and instructor in clinical investigation in the Krueger laboratory.
The researchers developed this tool by feeding 60 photos of cancerous melanomas and an equivalent batch of pictures of benign growths into image processing programs. They developed imaging biomarkers to precisely quantify visual features of the growths. Using computational methods, they generated a set of quantitative metrics that differed between the two groups of images—essentially identifying what visual aspects of the lesion mattered most in terms of malignancy—and gave each biomarker a malignancy rating.
By combining the data from each biomarker, they calculated the overall Q-score for each image, a value between zero and one in which a higher number indicates a higher probability of a lesion being a cancerous.
As previous studies have shown, the number of colors in a lesion turned out to be the most significant biomarker for determining malignancy. And some biomarkers were significant only if looked at in specific color channels—a finding the researchers say could potentially be exploited to identify additional biomarkers and further improve accuracy.
“I think this technology could help detect the disease earlier, which could save lives, and avoid unnecessary biopsies too,” says Gareau. “Our next steps are to evaluate this method in larger studies, and take a closer look at how we can use specific color wavelengths to reveal aspects of the lesions that may be invisible to the human eye, but could still be useful in diagnosis.”
This work was supported in part by the National Institutes of Health and in part by the Paul and Irma Milstein Family Foundation and the American Skin Association.
 (opens in new window)Experimental Dermatology, online: December 19, 2016 (opens in new window)Experimental Dermatology, online: December 19, 2016Digital imaging biomarkers feed machine learning for melanoma screening(opens in new window) Daniel S. Gareau, Joel Correa da Rosa, Sarah Yagerman, John A. Carucci, Nicholas Gulati, Ferran Hueto, Jennifer L. DeFazio, Mayte Suárez-Fariñas, Ashfaq Marghoob and James G. Krueger |


