Structure shows how a key protein in gene activation is controlled
 (opens in new window)
(opens in new window)
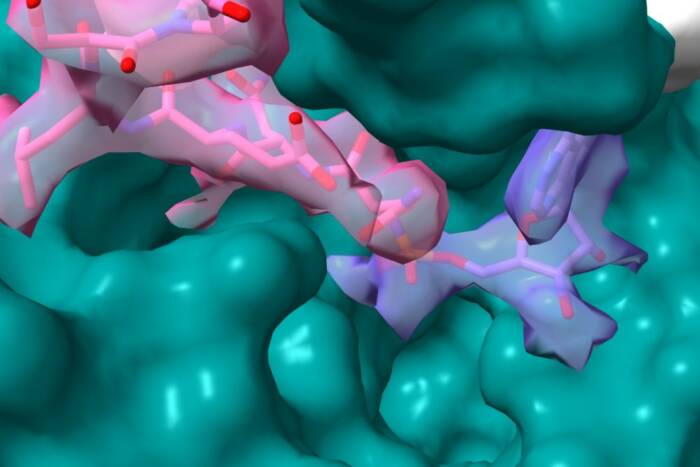
The expression of genes by RNA polymerase in bacteria depends on the presence of the σ protein. New research shows that when σ is not bound to DNA, its conformation resembles the structure it takes when bound to its inhibitor FlgM (above).
Having good genes is not enough; they each need to be expressed at the right time and place. By solving the structure of a protein called σ, researchers at Rockefeller University reveal a new mechanism by which bacteria prevent premature and precocious activation of their genes.
Bacteria use the σ protein to control when their genes are turned on and off. When σ binds to DNA the adjacent gene is turned on; when it’s not bound, that gene is turned off. A crystal structure depicting σ28, the initiation factor from the bacteriaAquifex aeolicus, bound to its inhibitor protein FlgM has previously shown that FlgM inhibits σ28 by packing together its DNA binding domains, which interferes with σ28’s ability to recognize DNA. FlgM also covers up the part of σ28 that interacts with the RNA polymerase, needed for gene expression. But Seth Darst, head of the Laboratory of Molecular Biophysics, and Margareta Sörenson, a former graduate student, thought that perhaps σ28 adopted the same structure even without FlgM, as a way to regulate itself.
“We found that free σ28, in solution, favors the conformation where all the DNA binding domains are tightly packed together,” says Darst. “But, unlike when it is bound to FlgM, the part of the protein that interacts with the RNA polymerase is still accessible. When σ28 binds to the RNA polymerase, this causes a conformational change in the σ28 protein, opening up its DNA binding regions and allowing it to bind to and activate the right genes at the right times.”
Proceedings of the National Academy of Sciences 103(45): 16722-16727 (November 7, 2006)(opens in new window)