An old antibiotic may combat drug-resistant tuberculosis
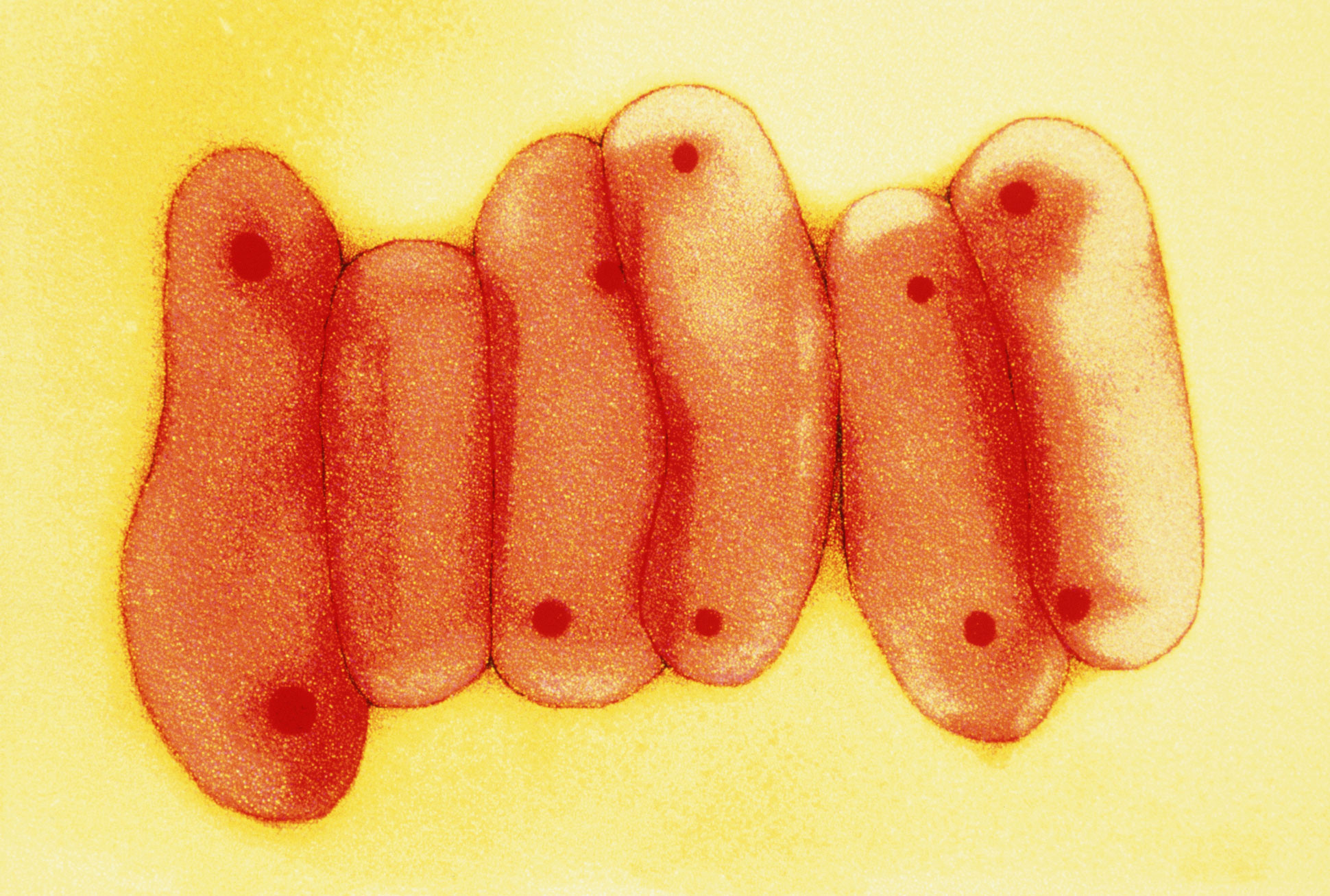
Mycobacterium tuberculosis, the microbe responsible for tuberculosis, often mutates in ways that make it resistant to antibiotics.
For decades, antibiotics have helped hold tuberculosis back, saving untold millions of lives. But their limits are being seriously tested as the bacterium’s tendency to mutate has led to a steady rise in drug-resistant strains. Without new drugs, scientists fear that tuberculosis, deemed largely a controllable disease, may not remain so.
In search of new antibiotics, Rockefeller scientists have landed on an existing compound, naturally produced by a bacterium. Their research, published in the Proceedings of the National Academy of Sciences(opens in new window), elucidates how sorangicin A, first discovered in the 1980s, can destroy even the antibiotic-resistant bacteria that cause tuberculosis.
The findings suggest that the compound may be a good candidate for further development as a first-line antibiotic for tuberculosis.
“Sorangicin inhibits regular strains in very much the same way as rifampin, one of the primary choices for tuberculosis antibiotics. But now we show that, through a different mechanism, it also traps those variants that escape rifampin,” says Elizabeth Campbell, a research associate professor at Rockefeller.
Dual action antibiotic
The antibiotic rifampin works by blocking RNA polymerase (RNAP), an enzyme crucial to bacteria’s survival. This enzyme walks along a strand of DNA, using the genetic information to build an RNA molecule, one nucleotide at a time. Rifampin snaps into the cavity of one of RNAP’s pockets, and physically clogs the path of the RNA molecule when it’s no more than two or three nucleotides long. Without the proper RNA blueprints, the bacterium can’t make new proteins, and dies.
Bacteria resistant to rifampin have mutations that change the structure of that rifampin-friendly pocket in RNAP. To find an alternative drug for these strains, researchers have been looking for molecules that fit into other pockets of bacterial RNAP.
At first glance, sorangicin would not seem a likely candidate since it targets the same RNAP pocket as rifampin. But it appears to work anyway: a few years ago, Campbell and others discovered that some of the strains that had developed resistance to rifampin were still vulnerable to sorangicin.
Flexible and early
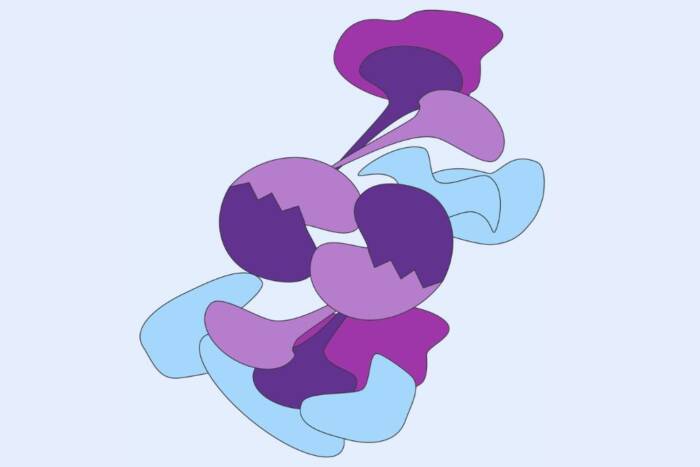
In the new study, the researchers used crystallography to visualize the interactions between sorangicin and RNAP’s pocket in mutant bacteria. They found that the very mutation that repels rifampin from the pocket also creates a flexible molecular lid on the pocket, which moves to let sorangicin in.
The researchers then used cryo-electron microscopy to see what happens when sorangicin binds to RNAP. In regular, non-resistant bacteria, sorangicin cut the RNA production short, similar to rifampin. But in the mutant strains, sorangicin trapped RNAP at an unexpected stage of transcription, early on before the production of RNA molecules even begins. “This was surprising,” Campbell says. “It tells us to look at all stages of the process when designing new antibiotics.”
Many tuberculosis patients have other health problems requiring medication, such as HIV. Treatment for these patients can be complicated because rifampin reduces the effectiveness of other medications by speeding their metabolism. Lab studies revealed that sorangicin has a milder effect on the enzymes involved in drug metabolism. “If sorangicin can be developed into a medication, it might be especially helpful for people with co-morbidities,” Campbell says.