Activation of tumor suppressor gene p53 much more complex than previously believed
It’s the biochemist’s twist on the old light bulb jokes: how many proteins does it take to activate a gene?
Scientists in Robert Roeder’s Laboratory of Molecular Biology and Biochemistry at Rockefeller University now know that, at least for gene activation by the tumor suppressor p53, the answer is as many as five — and perhaps more — proteins for a single early step in this process. Reporting in the June 11 issue of the journal Cell, Roeder, with first author Woojin An and graduate student Jaehoon Kim, also provides the first direct evidence that chemical changes to DNA packaging proteins called histones regulate transcription, or activation, of p53 and other target genes, a finding that has major implications for the treatment of human diseases, including cancer.
“There have been many studies correlating histone modifications and transcriptional regulation, but none of the studies directly showed that histone modification itself actually regulates transcription,” says An. “Our results show — as was generally assumed — that chemical modifications of histones indeed are required for the activation of gene transcription.”
“The regulation of gene expression is one of the most important and actively studied areas in biology today, and an understanding of the central role of histone modifications in gene activation has enormous implications for understanding both normal and abnormal cellular processes,” adds Roeder, last year’s recipient of the Albert Lasker Award for Basic Medical Research. “The p53 protein is an important tumor suppressor, and by looking at key target genes, we have found that there is a surprising complexity to p53-mediated gene activation, which occurs as a direct result of the function of several histone-modifying enzymes.”
The p53 gene is mutated in many cancers, and scientists are trying to determine the consequences of this abnormality and also how the p53 gene product, called a tumor suppressor, normally acts in the human body to monitor and to avoid harmful consequences of damaged DNA. When p53 is activated in response to DNA damage, it binds to and activates so-called target genes, which produce proteins that execute one of two possible responses. These proteins either arrest cell growth so that the cell can repair the DNA damage and grow normally or they kill the cell before it can generate a tumor. Scientists still do not properly understand the conditions that influence one DNA-damage response over the other.
In 1997, Roeder and Wei Gu, then a postdoc in Roeder’s lab, showed that an enzyme called p300/CBP, known to modify the chemical composition of histone tails, serves as a transcriptional coactivator for p53. Coactivators are regulatory proteins that, together with activator proteins, are required to turn genes on.
But Roeder and Gu also showed that p300 could chemically modify and alter the function of p53 itself, thus showing for the first time, as is now commonly observed, that histone-modifying enzymes can also modify regulatory factors, such as p53. This finding raised questions regarding the functions of the enzymes that are most important for transcription.
Scientists believe that histone modifications are crucial actors in the activation and repression of gene expression. Histones help to package DNA, the hereditary material of life, into each cell’s nucleus. The double-helical strand of DNA wraps around a ball of histones consisting of four distinct proteins: H2A, H2B, H3 and H4. This fundamental unit, called a nucleosome, is repeated at regular intervals throughout the length of DNA and, under a microscope, resembles “beads on a string.” Strings of nucleosomes coil up further to form a more compact chromatin and even further to become the familiar X-shaped chromosomes of human cells.
Beginning with early observations by Rockefeller University’s Vincent Allfrey, research by Rockefeller University scientist C. David Allis and others has provided a large body of evidence that correlated chemical modifications at specific locations on the tails of histone proteins with gene activation.
In 2002, An, Roeder and their Rockefeller colleagues provided important new information about the role of histone tails in gene activation. Scientists knew that histone tails repressed gene activation by preventing transcription factors — proteins that help read out the information encoded in DNA — from gaining access to DNA. An and colleagues, using a test tube system of coiled up chromatin created from engineered or recombinant histones and DNA, showed that — as Allfrey and Allis had predicted — histone tails and associated modifications are required for reversing the repression of transcription and that p300 plays an important role in this process as a histone-modifying enzyme.
Scientists elsewhere had previously shown that two other histone-modifying enzymes, called CARM1 and PRMT1, worked with p300 to mediate the function of a class of transcriptional regulatory proteins called nuclear hormone receptors. And like p300, CARM1 and PRMT1 can modify both histones and regulatory factors.
To determine if CARM1 and PRMT1, alone or in conjunction with p300, also are involved in gene activation by p53, An and his co-workers put all these proteins in a test tube with pure p53 and tested their function on a target gene wrapped up in a chromatin structure with histones.
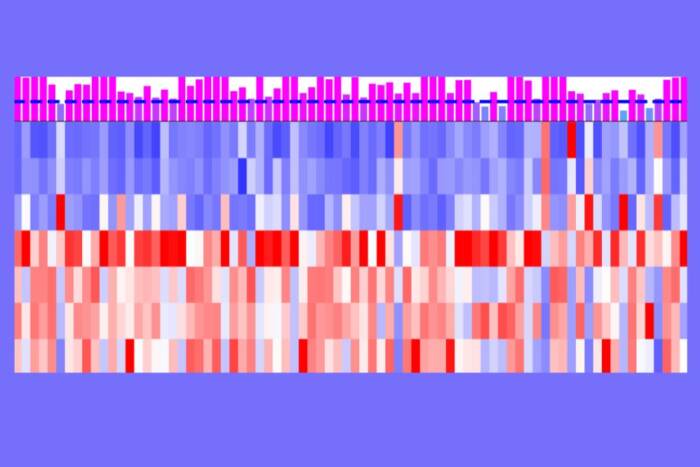
An found that the three proteins worked synergistically in mediating gene activation by p53. And surprisingly, An also observed that the proteins followed a specific order: activation of the p53 target gene GADD45 — a gene involved in the repair of damaged DNA — was strongest when An added PRMT1 first, p300 second and CARM1 third. Moreover, when analyzed with recombinant chromatin containing mutated histones that could not be modified, he failed to see gene activation by p53 — thus proving that histone modifications are indeed necessary for p53 function.
To show that the results he obtained in test tube studies also occur in living cells, An exposed cells to ultraviolet light, which inflicts DNA damage and causes GADD45 to be highly expressed, and watched the recruitment of cofactors. To do this he used an antibody-based assay called ChIP (chromatin immunoprecipitation), which enabled him to look at proteins sitting on the GADD45 gene in a living cell. Consistent with the test tube results, the first complexes to arrive at the scene, within two hours of exposure to UV light, were p300 and PRMT1. CARM1 showed up within four hours of UV irradiation. Somewhat surprisingly, the assay also showed p53-dependent accumulation of at least two other histone-modifying enzymes on the GADD45 gene during the activation process.
“It is surprising that so many proteins are required in the initial events — the histone modifications — leading to activation of a single gene,” says Roeder, the Arnold and Mabel Beckman Professor at Rockefeller University. “Now the stage is set to determine if all these proteins are used by all p53-responsive genes, or during all of the different stress responses that activate these genes, and how they influence the outcome, for example cell death vs. cell growth arrest, of the DNA-damage responses.”
This research was supported by a grant from the National Institutes of Health. Roeder is a member of the Pels Family Center for Biochemistry and Structural Biology at Rockefeller.