Parasites reveal how evolution has molded an ancient nuclear structure
Long before animals evolved from sponges, and before plants evolved out of algae, there was a pivotal event that allowed complex, multicellular organisms to arise: the development of the nucleus in single-celled organisms. Eukaryotes, one of the three main branches of living organisms, are defined by the presence of this nucleus and its surrounding membrane, which houses and protects our genetic material. All of the multitude of molecules that need to pass into or out of the nucleus do so via a channel-like structure embedded in this membrane—the nuclear pore complex.
Michael Rout, head of the Laboratory of Cellular and Structural Biology, and Brian Chait, head of the Laboratory of Mass Spectrometry and Gaseous Ion Chemistry, together with their colleagues, have been working to understand the nuclear pore complex for nearly two decades. They first isolated and described the components of the yeast nuclear pore complex in 2000 and then released a first draft of its structure in 2007. In yeast, as well as in humans, the nuclear pore complex is composed of an inner ring sandwiched between two outer rings: one facing in and one facing out. Attached to these outer rings are classes of proteins that are compatible with the unique chemistry of either the nucleus or cytoplasm found in the rest of the cell. This asymmetric distribution of nuclear pore components is important for helping to establish the directionality of transport through the pore.

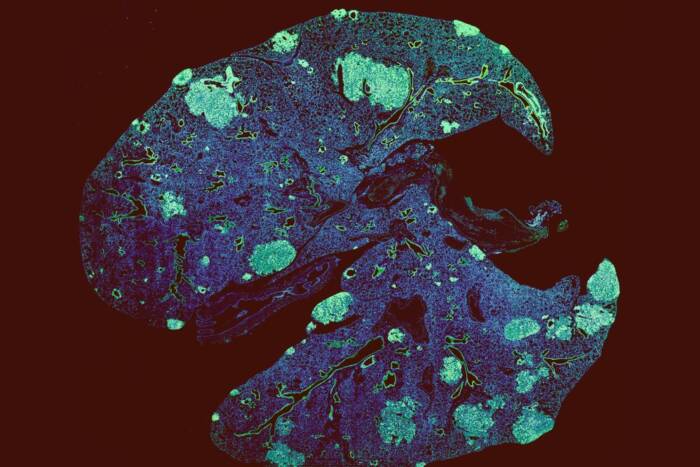
Pretty parasites: Trypanosoma brucei parasites, showing DNA (blue), and nuclear pore complexes (green). Unlike other eukaryotes, trypanosome nuclear pore complexes have a symmetrical distribution of proteins on their inside and outside faces.
The complex is so fundamental that its architecture was thought to be shared by all eukaryotes. However, Samson Obado, a research associate in the Rout lab, has found a eukaryotic species that has pores with a structure unlike any that has yet been studied, a finding which implies that the pore’s evolution is probably more involved than had previously been assumed. Obado’s results were published in February in PLoS Biology.
“We were driven by curiosity about the evolution of the nuclear pore complex,” says Obado. Almost everything we know about the complex is from yeast and humans, which although quite far apart in evolutionary terms are nevertheless closely related when compared to plants and many other single-celled eukaryotes. “But what did the original pores look like, and how have they developed in different eukaryotes?” Obado wondered.
He was the perfect candidate to investigate this, as his graduate studies focused on trypanosomes; these parasites, responsible for serious diseases such as sleeping sickness and Chagas disease, diverged from the families that developed into yeast, humans, plants, and other eukaryotes roughly one and a half billion years ago—close to the time of the last common ancestor of all eukaryotes.
“Unusual and quirky”
“Compared with many other eukaryotes, trypanosomes have an unusual and quirky molecular biology. A key example is how they transcribe their genes into messenger RNAs for translation into proteins, which is very different from textbook models. Furthermore, because they are so divergent, you can’t just search for gene sequences similar to those in yeast or humans,” Obado says.
Rout, Chait, and colleagues worked together with Mark Field of Dundee University in Scotland to identify putative nuclear pore components from trypanosomes. To do so, the team walked protein by protein through the nuclear pore complex, purifying each one along with other proximal proteins. These other proteins, they figured, could be part of the nuclear pore complex as well. Obado then purified the new proteins to determine whether they were also part of the complex, along with their proximal proteins, and so on until he had a complete survey of all the proteins involved. Finally, they worked with the university’s Electron Microscopy Resource Center to determine where key components are located with respect to each other. The result was a complete first picture of the entire trypanosome pore structure.
The teams’ results show that the architecture of the inner ring of the nuclear pore complex is fundamentally similar in trypanosomes, yeast, plants, and vertebrates, suggesting an ancient origin for this common feature. In contrast, the trypanosome nuclear pore complex possesses a unique mechanism tethering it to the surrounding nuclear envelope, and the outer ring is less well “conserved” by evolution, with additional never-before-seen components. However, the most notable difference between trypanosome nuclear pore complexes and others that have been examined to date, is that the trypanosome’s nuclear pore complex exhibits a near-complete symmetry of its components, and lacks almost all the proteins in yeast and humans that are required to establish an asymmetric assembly within the pore. Instead, it seems that soluble proteins in the nucleus and cytoplasm are responsible for setting transport’s directionality in these parasites for both proteins and RNA.
The fact that the trypanosome pore has such dissimilarities with the pore of humans may provide an opportunity, says Obado. “These differences may offer something we can target therapeutically without risking harm to our own transport mechanisms.”
Evolutionary Implications
These discoveries also lend further credence to an idea in evolutionary theory first suggested by Rout and his colleagues. The theory, known as the “protocoatomer hypothesis,” suggests that the nuclear pore and nuclear envelope share a common ancestor with structures that form coats on other membrane-bound structures in the cell, like those in the Golgi apparatus and endoplasmic reticulum. Indeed, the trypanosome nuclear pore components, despite their dissimilarities from other eukaryotes, still retain the kinds of structures and organization found in these other coats, the scientists say.
Moreover, since the inner core of the trypanosome nuclear pore complex is structurally well preserved and similar to those from other eukaryotes, Rout and his team believe this core represents the original units of a simpler nuclear pore complex that predates the last ancestor of all eukaryotes, and thus may provide new clues as to how the nucleus originally evolved.
 PLoS Biol 14(2): e1002365 PLoS Biol 14(2): e1002365Interactome Mapping Reveals the Evolutionary History of the Nuclear Pore Complex Samson O. Obado, Marc Brillantes, Kunihiro Uryu, Wenzhu Zhang, Natalia E. Ketaren, Brian T. Chait, Mark C. Field, and Michael P. Rout |