Researchers discover new antibiotics by sifting through the human microbiome
Most antibiotics in use today are based on natural molecules produced by bacteria—and given the rise of antibiotic resistance, there’s an urgent need to find more of them. Yet coaxing bacteria to produce new antibiotics is a tricky proposition. Most bacteria won’t grow in the lab. And even when they do, most of the genes that cause them to churn out molecules with antibiotic properties never get switched on.
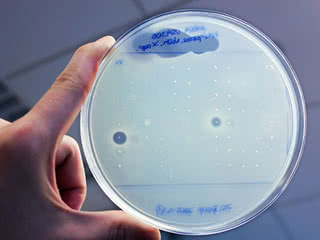
Toxic radius: The researchers placed tiny droplets of 25 newly discovered antibiotics on a carpet of beta-lactam resistant S. aureus. They identified two compounds that generated circles of dead bacteria (dark spots) around each droplet.
Researchers at The Rockefeller University have found a way around these problems, however. By using computational methods to identify which genes in a microbe’s genome ought to produce antibiotic compounds and then synthesizing those compounds themselves, they were able to discover two promising new antibiotics without having to culture a single bacterium.
The team, which was led by Sean Brady, head of the Laboratory of Genetically Encoded Small Molecules, began by trawling publicly available databases for the genomes of bacteria that reside in the human body. They then used specialized computer software to scan hundreds of those genomes for clusters of genes that were likely to produce molecules known as non-ribosomal peptides that form the basis of many antibiotics. They also used the software to predict the chemical structures of the molecules that the gene clusters ought to produce.
Unearthing the humimycins
The software initially identified 57 potentially useful gene clusters, which the researchers winnowed down to 30. Brady and his colleagues then used a method called solid-phase peptide synthesis to manufacture 25 different chemical compounds.
By testing those compounds against human pathogens, the researchers successfully identified two closely related antibiotics, which they dubbed humimycin A and humimycin B. Both are found in a family of bacteria called Rhodococcus—microbes that had never yielded anything resembling the humimycins when cultured using traditional laboratory techniques.
The humimycins proved especially effective against Staphylococcus and Streptococcus bacteria, which can cause dangerous infections in humans and tend to grow resistant to various antibiotics. Further experiments suggested that the humimycins work by inhibiting an enzyme that bacteria use to build their cell walls—and once that cell-wall building pathway is interrupted, the bacteria die.
A similar mode of action is employed by beta-lactams, a broad class of commonly prescribed antibiotics whose effect often wanes as bacteria develop ways to resist them. Yet the scientists found that one of the humimycins could be used to re-sensitize bacteria to beta-lactams that they had previously outsmarted.
Synergistic effects
In one experiment, they exposed beta-lactam resistant Staphylococcus microbes to humimycin A in combination with a beta-lactam antibiotic, and the bugs once again succumbed. Remarkably, that held true even when humimycin A had little effect by itself—a result that Brady attributes to the fact that both compounds work by interrupting different steps in the same biological pathway.
“It’s like taking a hose and pinching it in two spots,” he says. Even if neither kink halts the flow altogether on its own, “eventually, no more water comes through.”
To further test that proposition, Brady and his colleagues infected mice with a beta-lactam resistant strain of Staphylococcus aureus, a microbe that often causes antibiotic-resistant infections in hospital patients. Mice that were subsequently treated with a mixture containing both humimycin A and a beta-lactam antibiotic fared far better than those treated with only one drug or the other—a finding that could point towards a new treatment regimen for humans infected with beta-lactam resistant S. aureus.
Brady hopes that this discovery will inspire scientists to mine the genomes of bacteria for more molecules that could yield similarly useful results. And he looks forward to applying his methods to the many bacterial species that lie beyond the human microbiome, and that might harbor their own molecular treasures—not to mention the even greater number of bacteria whose genomes have not yet been sequenced, but that undoubtedly will be over time.
This research was supported by the National Institutes of Health (grants U19AI109713 and F3229AI110029) and the Rainin Foundation.
 Nat Chem Biol. 2016 Oct 17. doi: 10.1038/nchembio.2207. Nat Chem Biol. 2016 Oct 17. doi: 10.1038/nchembio.2207.Discovery of MRSA active antibiotics using primary sequence from the human microbiome. Chu J, Vila-Farres X, Inoyama D, Ternei M, Cohen LJ, Gordon EA, Reddy BV, Charlop-Powers Z, Zebroski HA, Gallardo-Macias R, Jaskowski M, Satish S, Park S, Perlin DS, Freundlich JS, Brady SF |