Scientists map the network of SARS-CoV-2’s helpers inside human cells
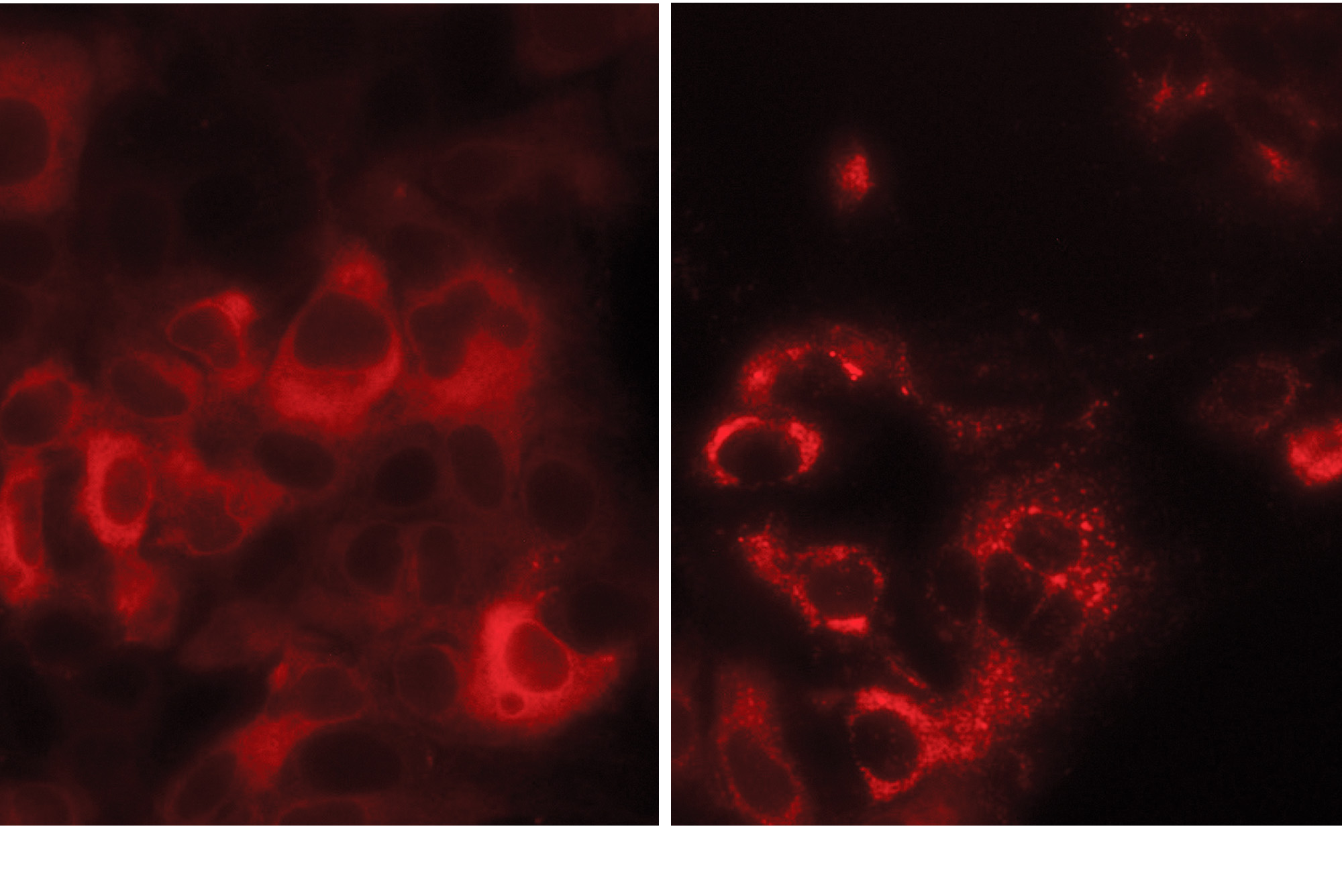
In this cell cluster, TMEM41B proteins (red) are normally diffused throughout each cell (left). But upon SARS-CoV-2 infection, they redistribute to form large clumps (right).
Like all viruses, SARS-CoV-2 is in the grey zone between a chemical and a living being. Its biological machinery is too scant to propagate on its own, and instead the virus hijacks proteins of the host to generate new copies of itself. Scientists hope that by developing drugs that disrupt such proteins, it might become possible to deal a fatal blow to the virus.
Now a new study(opens in new window) maps out this intricate network of helpers for SARS-CoV-2 and three other coronaviruses that cause the common cold. The results identify over a hundred human proteins without which these viruses cannot replicate inside the cells and cause disease.
Among them is a protein that stands out because it is an absolute requirement for infection by the four coronaviruses. Moreover, it is also needed by viruses from an entirely different family, flaviviruses, including those that cause yellow fever, West Nile, and Zika disease, according to a second study(opens in new window).
“The fact that this protein is important to multiple different viruses makes it a promising and high-priority target for drug development efforts,” says Rockefeller virologist Charles M. Rice, who co-led the studies with John Poirier at New York University. Both studies are published in Cell.
Shared vulnerabilities
Once it has entered a host cell, SARS-CoV-2 co-opts hundreds of molecules to make copies of its RNA and wrap them up as thousands of new viruses.
The researchers set out to identify these human proteins, both those that are specific to SARS-CoV-2, and those shared between several coronaviruses. “We need to focus not only on the current pandemic but also think about the future,” says William Schneider, a postdoc in Rice’s Lab(opens in new window). “If multiple members of the coronavirus family require the same factor, it’s likely that an unknown coronavirus emerging in the future will also require it.”
Casting their net as wide as possible, the researchers studied the effects of about 19,000 genes, nearly the whole genetic repertoire of human cells, on the viruses’ ability to replicate. They used the gene-editing tool CRISPR to inactivate only one of the genes in any given cell, and subjected the cells to the viruses.
Over the next couple of days, the viruses killed off massive numbers of cells. The few cells that survived the viral attack were those in which a critical gene had been disabled.
The results revealed an extensive network of human proteins and cellular pathways used by the four coronaviruses to complete their life cycle. “We can see some pathways, like those synthesizing cholesterol, is shared by all four, whereas others seem unique to only one of two of the coronaviruses,” says Hans-Heinrich Hoffmann, a postdoc in Rice’s lab. “Altogether, this catalogue is a rich resource to for exploring new drug targets for COVID-19 and potential future coronavirus outbreaks.”
Surprise protein
The genome-wide study also revealed that coronaviruses rely on several proteins whose functions are not fully understood. Among them was TMEM41B, a protein the team had encountered before.
In 2016, the researchers carried out a similar study for Zika virus, one of the major public-health concerns at the time, and found it required TMEM41B. Subsequent studies showed none of the other flaviviruses tested could replicate in cells missing this protein, but viruses from 12 other families were not impacted.
The researchers assumed TMEM41B was required for flavivirus replication only, until SARS-CoV-2 emerged. “Coronaviruses were some of the few viruses we hadn’t tested yet, and to our surprise they also rely on TMEM41B,” Hoffmann says.
Recent studies into the cellular function of TMEM41B suggest it plays a role in autophagy, the process by which the cell digests and discards cellular debris. TMEM41B is involved in this process by helping wrap the debris in a fatty double membrane. Based on their lab experiments, the researchers suspect that viruses hijack TMEM41B to help wrap themselves in secure nooks inside the cell membranes, where they can hide and safely replicate their genetic material.
Remarkably, nearly 20 percent of East Asian populations have a variant of TMEM41B. In lab experiments, cells with this variant were less susceptible to flavivirus infection than cells with the more common form of TMEM41B. More research is needed to know whether this variant also offers protection against SARS-CoV-2.
The team plans to further investigate whether it’s possible to design a drug targeting TMEM41B in a way that allows it to continue doing its normal job, but impairs its ability to support a virus infection.