New algorithm cleans microbiome data with unprecedented efficiency
None of us are born with a fully functioning immune system, and the first few months of life are crucial for establishing strong lifelong defenses. Better understanding how germs influence the development of human immunology would reap innumerable benefits, but for years, scientists haven’t been able to answer the most basic question: where does inoculation start, in utero or at the moment we first encounter the world?
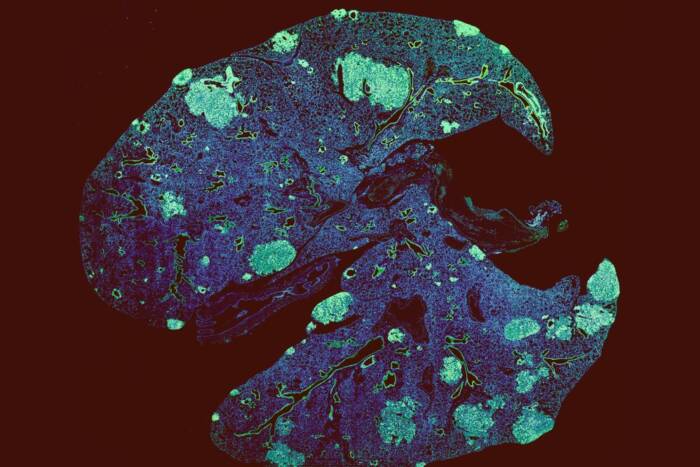
On the surface, it seems there couldn’t be an easier way to find out—just place a slice of the placenta under a microscope and look for a community of microbes. Yet no fewer than six papers on the topic have been published in top journals since 2014, bringing us no closer to a definitive answer: three show microbes; three show nothing.
Contamination is the problem. Although scientists can detect the most minute traces of microbial genetic information in a sample, distinguishing native bacteria and viruses from contaminants that ride into the experiment on skin, reagents, and lab equipment has proven difficult—so difficult that it’s still unclear whether the microbes that some studies have found in placentas were there before technicians worked with them. A recent study in Nature Biotechnology may have the key to solving this conundrum: a novel algorithm called SCRuB that allows researchers to remove contamination from microbiome data with unprecedented efficiency.
“Our key insight was that, instead of trying to identify whether a specific microbe is a contaminant, our algorithm models the entire contamination source,” says Liat Shenhav, an independent research fellow at Rockefeller’s Center for Studies in Physics and Biology who led the project. By taking a nuanced approach to decontaminating samples, the method allows scientists to make better use of so-called negative controls—blank samples processed along with the test sample—to remove decontamination. It is highly specific, and performs particularly well with microbes that may exist as both contaminants and native residents. “SCRuB outperforms current state-of-the-art methods by an average of 15-20 times.”
On the heels of their success, its developers wondered whether SCRuB could become widely applicable to microbiome research. For instance, they showed that predicting whether melanoma patients will respond to immunotherapy hinges on an accurate analysis of the microbes living in each unique tumor, enabled by specific decontamination. The team also showed that their algorithm can classify melanoma patients better than existing methods.
“Our understanding of microbial communities is just scratching the surface,” says Tal Korem of Columbia University, who co-led the study with Shenhav.” The field is generating data from diverse body sites and at a staggering pace. All of these studies could benefit from accurate decontamination using our method.”