New Mtb study offers a novel paradigm for understanding bacterial transcription
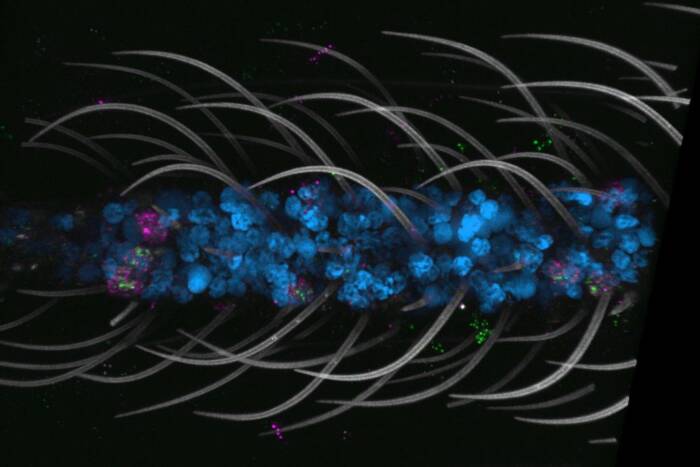
Researchers have discovered that the transcriptome of Mycobacterium tuberculosis mainly consists of incomplete transcripts resulting from pausing of the RNA polymerase on genomic DNA. (Credit: DrawImpacts)
The bacterium behind tuberculosis is a wily foe, adept at bobbing and weaving around the immune system and antibiotics alike. Mycobacterium tuberculosis (Mtb) has been notoriously difficult to eradicate, often dormant in the body for years only to reactivate when the time is ripe.
Now, new research reveals how Mtb controls its gene expression, which may provide clues as to how it adapts to challenging environmental conditions. The findings, published in (opens in new window)Nature, could ultimately offer drug targets that would stop Mtb in its tracks. “It’s a very smart bacterium, with a lot of tricks,” says Rockefeller’s Shixin Liu. “Now that we have exposed how it regulates gene expression, we can use that information to think about how we might inhibit its lifecycle.”
An enabling technology
Tuberculosis is the leading cause of death among infectious diseases worldwide, in no small part because Mtb possesses a remarkable repertoire of ways to regulate the transcription of RNA from DNA, which is subsequently translated into functional proteins. This flexibility enables the bacterium to adapt to changing environments and antibiotics in a human host. Liu and colleagues realized that the more we understood about how Mtb transcribes its genetic code, the more likely we would be able to eventually out-maneuver the pathogen. “A detailed characterization of the Mtb transcriptome is key to more effectively treating it,” Liu says.
Existing knowledge of the transcriptional machinery of bacteria, however, was largely derived from research on E. coli, a poor model for Mtb. Empowered by SEnd-seq, a transcriptomic profiling tool developed by Liu and senior research associate Xiangwu Ju that captures both 5’ and 3’ ends of RNA transcripts at once, the Liu lab teamed up with Rockefeller’s Mtb experts Jeremy Rock and Elizabeth Campbell to put the technology to use in characterizing the Mtb transcriptome.
“SEnd-seq had already generated new insights into E. coli, in previous work,” Liu says. “It provides greater resolution than standard RNA sequencing techniques.”
A universal pause
It wasn’t long before the collaboration— supported by the Stavros Niarchos Foundation Institute for Global Infectious Disease Research at Rockefeller— started generating crucial insights. Using SEnd-seq, Ju found that most Mtb transcripts were incomplete, comprised of short RNA molecules that stop well before the gene’s end—an unprecedented discovery for a bacterium.
The team then demonstrated that these short RNAs were not readily released from the chromosome to become free fragments. Rather, they largely remained bound to DNA and the enzyme RNA polymerase (RNAP). During transcription, RNAP and its associated sigma factor were working in fits and starts, routinely pausing as they transcribed Mtb’s genes. RNAP pausing is common in eukaryotes, which utilize the pauses as “breathers” during which the cell can re-evaluate whether changing conditions call for a different transcriptional plan.
But bacteria weren’t thought capable of such strategic pausing. “The findings were so unexpected that I initially didn’t believe them,” Rock says. “We can now begin to explore whether similar mechanisms are utilized in other bacterial taxa.”
The researchers suspect that RNAP pausing underpins Mtb’s ability to dynamically adjust gene expression in response to threats. If that’s true, then perturbing RNAP pausing could leave Mtb vulnerable. Since RNAP is a prominent TB drug target, new understanding of the polymerase’s pausing mechanism could lead to innovative pharmaceutical avenues.
“What we discovered in Mtb was completely unknown before,” Rock says. “But that’s just the beginning. Now our goal is to learn more about it and, hopefully, find a way to interfere with it.”