Study reveals how unexpected shifts in cell populations are revising our understanding of the aging process
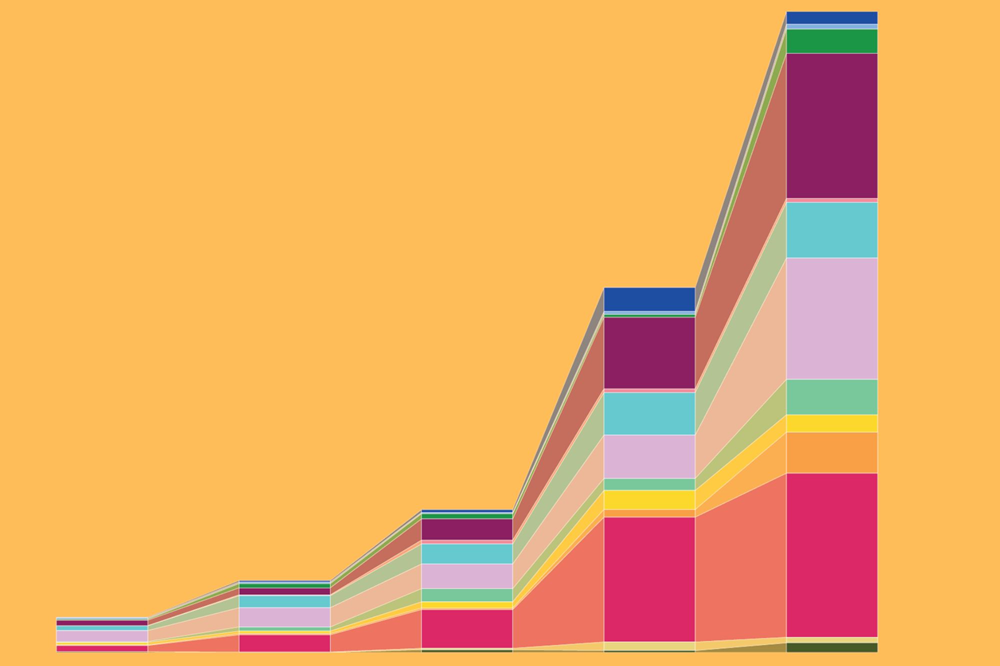
A chart showing the explosive expansion in proportion of Granzyme K+ CD8+ T cells in mice across multiple organs as they age (Cao lab)
If you looked at two snapshots of the same maple tree taken in July and December, you’d see a dramatic change from summer’s full green crown to winter’s bare branches. What those two photos don’t show you, however, is how the change occurred—gradually or all at once? In truth, deciduous trees tend to hold out for an environmental signal—a change in light or temperature—and then shed all their leaves within just a week or two.
When it comes to aging, we may be more like these trees than we realized.
According to groundbreaking new work from Rockefeller’s Laboratory of Single-Cell Genomics and Population Dynamics(opens in new window), mammals follow a similar aging trajectory at the cellular level. As described in a new paper(opens in new window) in Science, lab head Junyue Cao and his colleagues used single-cell sequencing to simultaneously scan more than 21 million cells from every major mouse organ across five stages of life. This enormous collection is now the world’s largest cellular atlas within a single study.
Their findings reveal that certain cell populations change in every organ, both in the same way and at the same time, during specific stages of life. This suggests that aging is not a linear process(opens in new window) but instead a developmental stage sparked by specific molecular cues.
“Some cells greatly expand in number while others drop off, and the cells that go through such changes are different depending on age,” says Cao. “What’s more, some of these changes are controlled by the same molecular features, so we might be able to target them to delay or even reprogram the aging process itself.”
From bespoke technique to universal platform
Single-cell sequencing, a specialty of Cao’s lab, is a method of genetic analysis that homes in on the genetic expression and molecular dynamics of individual cells, simultaneously revealing the identity of every cell studied. His team has previously used single-cell sequencing to discover new rare brain cell types and to track how brain cells age, among other purposes.
In the brain study, they also found different cell populations and cellular dynamics specific to different ages. For the current study, led by graduate student Zehao Zhang, they were curious to see if similar changes occurred elsewhere in the body.
To do that, Zhang adapted EasySci, a single-cell sequencing method developed by the group and used in their aging-brain study, to expand its cellular reach to include all the major organs of a mouse—a daunting task that Zhang managed single handedly.
“The most challenging aspect was optimizing EasySci to work across diverse mammalian organs while maintaining high-quality data,” he says. “Since most previous studies often focused on specific organs or used different protocols for different organs, developing a universal protocol required testing thousands of conditions across different organs before we could even begin the real experiment.”
As a result of Zhang’s efforts, EasySci is now a unified profiling platform for major mammalian organs, capable of systematically dissecting aging and disease mechanisms across an entire organism.
For the current study, they used it to reveal the single-nucleus transcriptome profiles of some 21 million cells taken from more than 600 samples of both male and female mice at five different life stages, from young to elderly.
Critical time windows
The team discovered more than 10 main cell types and 200 cell subtypes that consistently undergo significant age-associated depletion or expansion.
In early adulthood, for example (3 to 12 months in mice), specific cell subtypes within fat, muscle, and epithelial tissues saw a marked drop-off in number, while in advanced adulthood (12 to 23 months in mice), different types of immune cells explode in number.
Intriguingly, many of these changes were linked with specific gene expressions by the cells, regardless of where they were found. “We identified cell subtypes in different organs, where they may have different functions,” Cao says. “But they seem to be controlled by the same molecular process.”
“We’ve essentially identified the cellular basis of each phase change, and documented that they don’t happen gradually over time, but at specific stages of life,” he adds. “Now that we’ve identified the critical time windows that show very strong changes in distinct cell populations, this provides us important clues about how to intervene in the aging process.”
Immune cells in particular showed population booms in later life. “We found many different B cell and T cell subtypes that get strongly expanded in different organs,” Cao says. Excesses of these cells are known to cause inflammatory and autoimmune conditions. In fact, when the researchers examined two immunodeficient mice lacking such cells, they found that the depletion of B cells and T cells reversed changes in several other cell types associated with aging, highlighting cell regulatory networks in the aging process, Cao says.
They also found extraordinarily small clusters of new cell types, some numbering as few as 500 cells. What role they play in aging remains to be studied, but some extremely rare cell types have been shown to orchestrate critical functions, Cao says. “Take pituitary gland cells: this very small population secretes important hormones essential to growth, reproductive development, and organ function.”
Age and sex differences
Unexpectedly, they also found hundreds of cellular states that differed between male and female mice in every organ, Zhang says. These include adipocyte progenitor cells, which exhibit distinct molecular states between males and females, as well as a female-specific expansion of aging-associated B cells.
Put together, these age and sex differences may help explain why women, and especially older women, suffer from autoimmune conditions at a higher rate than men do.
They also underscore the importance of having sex-balanced cell samples in aging and disease studies, Zhang says. “Many studies focus on one sex to reduce costs and maintain consistency, but this finding highlights the importance of including both sexes to uncover generalized mechanisms or to develop sex-specific treatments.”
A goldmine for future research
The study’s 21-million-cell dataset, called PanSci, represents the largest single-cell-sequencing atlas of mammalian aging ever created, and Cao’s lab is already planning several future projects based on this resource. For instance, they intend to look deeper into the hundreds of cellular subtypes that show robust differences between male and female mice, as well as those involved in the aging process, many of which remain poorly characterized or studied.
“I think our findings could potentially be used to identify the cellular basis for some sex-specific diseases,” Cao says.
Scientists around the world are also welcome to mine PanSci for their own research, Zhang adds. “Researchers studying specific organs can extract organ-specific data, while those focusing on specific cell lineages such as immune or endothelial cells can extract the same cell types from different organs,” he says. “And because the dataset is well-curated and annotated, it’s ideal for training large machine-learning models for applications like age prediction, finding rare cell types, and building virtual cells for in silico perturbation studies.”




