Rockefeller researchers identify new mechanism that ensures accurate partitioning of genetic material in dividing cells

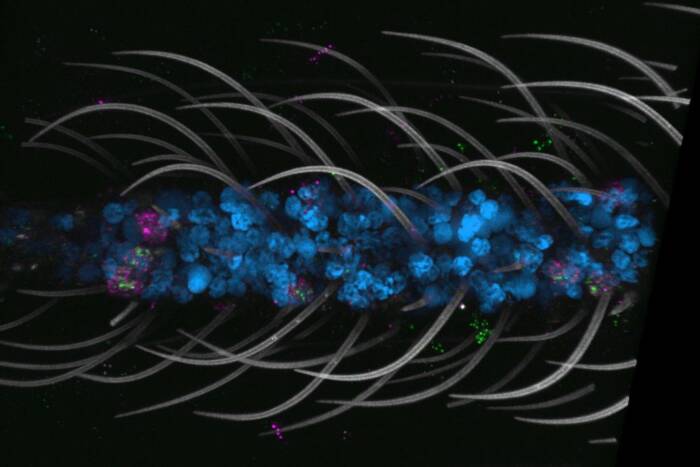
Centrosomes (orange) in a marsupial cell begin to move apart and form a bipolar spindle after the mitotic inhibitor monastrol has been washed away. Assistant Professor Tarun Kapoor and graduate fellow Lily Copenagle have identified a new mechanism by which microtubules (green) act to correctly align chromosomes (blue) during cell division. Reproduced from theJournal of Cell Biology, 2003, Volume 160, p. 678, by permission of The Rockefeller University Press.
Every minute, the human body replaces 300 million of its dying cells with new ones through the vital biological process known as cell division. When dividing and multiplying, a “parent” cell must segregate with exquisite precision each of its 46 chromosomes so that two “daughter” cells inherit all of its genetic information.
Mistakes in this process, called mitosis, can result in disease and developmental defects.
Using organic chemistry and state-of-the art live-cell imaging techniques, Assistant Professor Tarun Kapoor and his colleagues have identified a new mechanism in marsupial and human cell lines that contributes to the accurate alignment and segregation of chromosomes in vertebrate cells during mitosis.
These findings, reported in the current issue of theJournal of Cell Biology (March 3) by Kapoor, Rockefeller graduate fellow Lily Copenagle, and scientists at the Wadsworth Center in Albany, NY, and the Dartmouth Medical School, add to our understanding of the mechanisms by which cells ensure the accurate transmission of genetic information during cell division.
“This new mechanism reveals how chromosomes in a cell are aligned in the mitotic apparatus to be accurately segregated into two daughter cells,” says Kapoor, who heads the Laboratory of Chemistry and Cell Biology.
During mitosis, rod-like protein complexes in the cell called microtubules capture the duplicated chromosome and, along with other proteins, properly align it in preparation for its separation into daughter cells. A pair of duplicated chromosomes is properly aligned when it has one and only one attachment to each pole, called the centrosome.

A "textbook" model of the mitotic spindle. Rockefeller researchers have shown that the minus ends of microtubules provide a mechanism, along with a previously identified complementary mechanism mediated by the plus ends, for aligning and segregating chromosomes in the dividing cell.
Microtubules have polarity — a plus and a minus end — which results from the asymmetric assembly of the tubulin molecules that compose the microtubules. In mitosis, the minus ends of microtubules are usually associated with the centrosome, and the plus ends are associated with kinetochores, the plate-like structures on the duplicated chromosomes where microtubules attach.
Research by other scientists during the last 20 years has produced a “textbook” model called “search and capture” in which the plus ends of the microtubules capture the duplicated chromosomes. The new research by Kapoor and Copenagle reveals that the minus ends, previously considered to be static and undynamic, are actually very dynamic and provide a complementary mechanism for aligning and segregating chromosomes in the dividing cell.
“We needed to perturb the dividing cell in order to observe this minus-end capture mechanism,” says Kapoor. “But now that we know what it looks like, we see it happening in normally dividing cells.”
To perturb the dividing cell, Kapoor and Copenagle used a small molecule inhibitor called monastrol. While at Harvard Medical School, Kapoor and colleagues found that monastrol blocks the function of a key microtubule motor protein called Eg5, which is necessary for forming a bipolar spindle. Scientists hypothesize that Eg5 helps to separate the poles of the spindle. When dividing cells are exposed to monastrol, Eg5 is inactive and the spindle poles collapse, forming a monopolar spindle where both centrosomes lie in the center of an aster of microtubules. Removing monastrol results in a rapid reversal of this affect and the spindle reverts to its usual bipolar configuration.
“The general question we wanted to answer was: what happens if a chromosome is poorly oriented, such that its kinetochore cannot encounter the microtubules emanating from the centrosomes?” says Copenagle.
To answer this question, the researchers combined the powerful tools of small molecule inhibitors and live-cell microscopy.
“By using monastrol, we wanted to increase the probability of finding a chromosome that is not correctly attached; that is, a chromosome with an attachment to only one pole,” says Copenagle.
Using a spinning disk confocal microscope, which can image the dynamics of fluorescent proteins and chromosomes in a cell in real time, Kapoor and Copenagle observed dividing cells under the influence of monastrol. Real-time imaging of the cells was done in collaboration with Rockefeller’s Bio-Imaging Resource Center, directed by Alison North.
“The spinning disk confocal microscope produces the highest resolution images of a cell we have seen,” Kapoor says. “We can watch how a single chromosome in a vertebrate cell attaches itself to the mitotic apparatus.”
What they found was surprising: microtubules extended away from mono-oriented kinetochores out into the cytoplasm and looped back toward the spindle.
“We believe this looping and capture of microtubules occurs when the unattached kinetochore on a chromosome with only one attachment to a pole does not interact with microtubules from the opposite pole,” says Copenagle.
Importantly, the looping and capture of minus-ends of microtubules was also observed in cells that were not treated with monastrol.
Among the Rockefeller team’s findings was that chromosomes, and not just the centrosomes at poles, may directly contribute to the formation of microtubules that connect chromosomes to the mitotic apparatus.
“Our studies reveal the role centrosomes play in setting up the axis of cell division. The correct orientation of the cell division axis is essential for accurate development of multicellular organisms,” says Kapoor.
The researchers also found that the protein NuMA (Nuclear Mitotic Apparatus) assists in the looping and capture of the minus ends of microtubules. NuMA is known to aid in “bundling” microtubules at the poles and to interact with microtubule motor proteins. When NuMA was inhibited in human cells, the microtubules that would normally loop back remained extended in the cytoplasm.
“We now know that this new mechanism of loop and capture of microtubule minus ends, along with the search and capture of microtubule plus end, accounts for the attachment of all chromosomes to the spindle during mitosis,” says Kapoor.
In addition, these findings add to the body of knowledge Kapoor is compiling in his study of the mitotic protein Eg5.
“In my lab, we are using new potent cell-permeable small molecule inhibitors to validate Eg5 as a new target for anticancer therapy,” Kapoor says. “Understanding Eg5 inhibition helps us to clarify basic cellular mechanisms.”
This research was supported by the National Institute of General Medical Sciences, part of the federal government’s National Institutes of Health.