Special Proteins Transmit Signals Within Cells
Innovative laboratory technique reveals how messages influence cellular careers.
Special proteins play a key role in receiving and sending messages that influence the careers of healthy and diseased cells. In an innovative laboratory approach, Rockefeller University scientists developed probes that reveal the shape, structure and function of proteins that relay commands to govern a cell’s growth, role, movement and death.
Scientists know from previous studies that sections of signal-passing proteins, called src homology (SH) domains, help send messages within cells. If something damages the domains of the proteins and messages go awry, disease can occur.
“Experiments with the probes we developed provided a detailed structural picture of the SH domains in signaling proteins, which helps us to better understand the mechanics of how domains assist the proteins in transporting messages,” says David Cowburn, Ph.D., D.Sc., associate professor and head of the physical biochemistry laboratory(opens in new window) at Rockefeller. Cowburn and his colleagues published their findings in the Nov. 10 Journal of Biological Chemistry.
The communication process is tricky to study because many of the steps to pass along the messages–a series of signal proteins using their domains and other parts to combine, modify and release other proteins–are transient. Also, techniques that reveal the atomic and molecular structures of proteins, such as nuclear magnetic resonance spectroscopy (NMR) and X-ray crystallography, may not be able to observe the critical steps of the process. Cowburn’s creation and use of new probes addresses both problems.
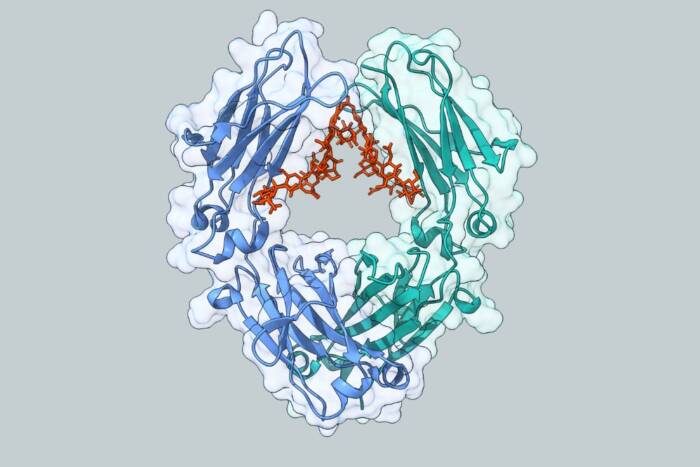
“With information we gleaned from the probe studies, we intend to create highly specific materials that trap or selectively interfere with the action of the domains and the signal proteins,” explains Cowburn, who has begun such investigations. “Results from these experiments may also provide leads for diagnostic and therapeutic agents to use in many areas of disease development in which signaling through SH domains occurs, such as cancer, diabetes and autoimmune disease.” In the probe studies, Cowburn and his colleagues built several that could find and bind to two SH domains, SH2 and SH3. Both domains frequently occur in proteins involved in cancer, diabetes and autoimmune disease. For the experiments, the scientists used the versions of SH2 and SH3 found in an enzyme, Ableson kinase (Abl), which if mutated can cause a form of leukemia.
SH2 and SH3 domains usually occur next to each other but have different functions. Often, the first step of signal transmission inside a cell–the flicking of a molecular switch from off to on–is the addition of a highly-charged phosphate group to a tyrosine, one of the 20 amino acids that make proteins. As SH2 domains bind to phosphorylated tyrosines, they register that the molecular switch is tripped and move the signal further along in the cell. The function of SH3 domains is less well understood, although they are known to bind to short proteins rich in proline, another amino acid. In the case of Ableson kinase, if the enzyme is mutated, the SH2 domain is required to make the protein one that causes cancer, while the SH 3 domain tries to inhibit or regulate the enzyme. The normal function of Ableson kinase is not known.
Two constraints in studying SH2 and SH3 domains are that they have a range of individual specificities and they can permit large proteins to form many associations. Because such interactions are important in explaining the complete activities of the signal process, a detailed structural picture of the molecular organization of the SH2 and SH3 domains is a significant research goal, Cowburn notes.
In his laboratory, Cowburn and his collaborators at the University of Minnesota created a series of probes, known as bivalent consolidated ligands. The probes have several binding portions, separated by sections called linkers, capable of binding simultaneously with the SH2 and SH3 domains.
The consolidated ligands bind more tightly and specifically than probes designed for SH2s or SH3s alone. The binding also depended on the length of the linkers, with the optimum distance similar to that expected based on models of combined SH2 and SH3 domains.
“Our results indicate that the consolidated ligands are useful for probing the structural and functional activities of many multidomain proteins containing SH2 and SH3 domains,” Cowburn says.
The experiments also revealed the position of the probe ends in relation to the location of the SH domains and more information about the binding sites within the domains. For example, the investigators found that the binding sites can come moderately close to one another.
In related studies published in the Oct., 15 Structure, Cowburn and his colleagues used nuclear magnetic resonance techniques to determine the structure of the Ableson SH3 domain when the enzyme is in a solution. They determined the structure of the Abelson SH2 domain in 1993.
Cowburn’s coauthors on the Journal of Biological Chemistry study include Jie Zheng, Ph.D., postdoctoral associate, Qinghong Xu, B.S., graduate fellow, and George Barany(opens in new window), Ph.D., professor in the Department of Chemistry(opens in new window) at the University of Minnesota(opens in new window). The National Institute of General Medical Sciences(opens in new window), part of the federal National Institutes of Health(opens in new window), supported the research.