Researchers profile active genes in neurons based on connections
When it comes to the brain, wiring isn’t everything. Although neurobiologists often talk in electrical metaphors, the reality is that the brain is not nearly as simple as a series of wires and circuits. Unlike their copper counterparts, neurons can behave differently depending on the situation.
Researchers in Jeffrey Friedman’s Laboratory of Molecular Genetics have devised a way to create snapshots of gene expression in neurons based on their connections. These snapshots contain exhaustive lists of the active genes within neurons that send information to a synapse, the junction between neurons.

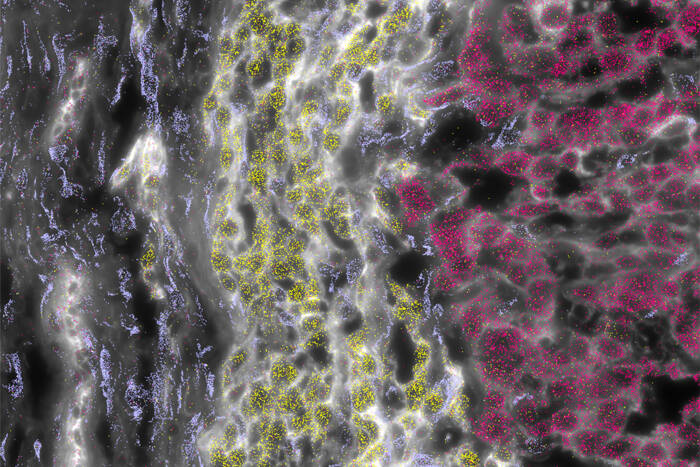
A new mouse TRAP: The team tested their profiling technique on midbrain mouse neurons (blue) that use the neurotransmitter dopamine to send signals to a brain region known as the nucleus accumbens. To do so, they tagged protein-assembling ribosomes (red) using a small antibody that bound a fluorescent protein (green).
Their new technique, called Retro-TRAP, merges two approaches to understanding the brain: mapping all of its connections and profiling gene expression within populations of neurons, Friedman says. “We hope that Retro-TRAP will be broadly used and provide a more granular understanding of how complex neural circuits function and ultimately lead to better treatments for neurological and neuropsychiatric disorders.”
“Refinements in neuroscience over time have allowed us to explore how the nervous system works in ever-greater detail, and the approach we have developed continues this trend,” says Mats Ekstrand, a research associate in the laboratory. “By building on existing techniques, we are now able to take a closer look at the types of cells involved in a particular circuit and what they are doing.”
In the long run, these sorts of insights might help explain why some diseases, such as Parkinson’s Disease, afflict particular sets of neurons, or someday make it possible to precisely target treatments at a dysfunctional neural circuit, rather than bathing the entire brain in drug.
The researchers modified a technique known as translating ribosome affinity purification (TRAP), developed at Rockefeller by Nathaniel Heintz, Paul Greengard and others to identify gene expression using green fluorescent protein to tag protein-assembling machines called ribosomes.
In research published today in Cell, Ekstrand, graduate student Alexander Nectow and colleagues describe how they used Retro-TRAP to introduce green fluorescent protein to the neuron via a virus that travels backwards from a synapse into the body of a mouse neuron. The researchers used a small antibody to link the ribosome with the fluorescent protein. Then, using these fluorescent tags, the researchers pulled out the ribosomes and sequenced the genetic messages passing through them. In this way, they produced a list of active genes.
To test their technique, the team focused on inputs to a well-studied part of the brain, the nucleus accumbens, which integrates information from throughout the brain, including regions involved in executive function, memory, depression, reward-related behavior, feeding and other functions, Nectow says.
“We wanted to target a selected number of inputs into the nucleus accumbens because we figured we might be able to get some molecular clues as to why it is important in regulating so many functions,” Nectow says.
Using Retro-TRAP, they created molecular profiles of neurons extending from the hypothalamus and ventral midbrain that project to the nucleus accumbens. The results confirmed that Retro-TRAP works.
“The nucleus accumbens receives a lot of signals from the ventral midbrain via the neurotransmitter dopamine, and, as expected, the genes we sequenced included many associated with dopamine neurons,” Nectow says.
Their data also contained some new discoveries. For instance, they found some neurons in the lateral hypothalamus express the p11 gene implicated in depression. After some further work, they found these neurons also tended to express a protein called orexin, a regulator of sleep and feeding — suggesting a molecular association between depression and some of its symptoms.
Retro-TRAP merges two approaches to understanding the brain: mapping all of the connections within it and profiling gene expression within populations of neurons, Friedman says. “We hope that Retro-TRAP will be broadly used and provide a more granular understanding of how complex neural circuits function and ultimately lead to better treatments for neurological and neuropsychiatric disorders.”
 |
Cell 157, 1230–1242 (May 22, 2014) Molecular Profiling of Neurons Based on Connectivity Mats I. Ekstrand, Alexander R. Nectow, Zachary A. Knight, Kaamashri N. Latcha, Lisa E. Pomeranz and Jeffrey M. Friedman |