Untargeted Analysis of Lipids
Un-targeted, or hypothesis generating, analysis of lipids is carried by combining high mass accuracy measurement of precursors with high mass accuracy fragmentation data. Molecules are most often selected based on abundance where after a selected mass is excluded for a chromatographic relevant time period (Data Dependent Acquisition, DDA, or ‘shotgun analysis’). Liupids analytes are separated using reversed phase (RP) chromatography, often in the form of C18 particles. Currently, the Center using two Data Dependent Acquisition strategies: 1) a pooled sample, composed of each of the samples to be analyzed, is analyzed extensively to create a library of identified lipids. Individual samples are then analyzed in polarity switch mode and pool-identified analytes are mapped to MS1 signals of the samples, or, 2) Each sample is analyzed separately in positive and negative mode and fragmentation data are collected using DDA.
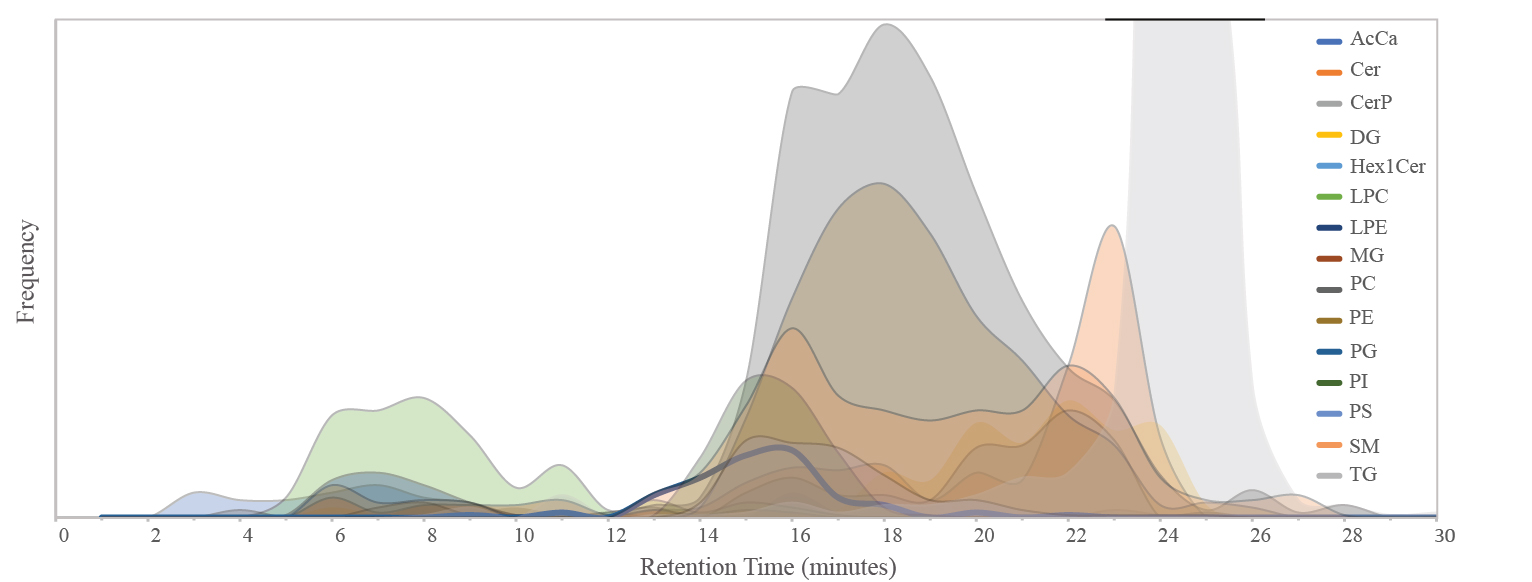
Data are analyzed using LipidSearch. Even high mass accuracy/high resolution fragmentation data can be difficult to match to the correct lipid because of identical or poor fragmentation. However, most lipid groups has unique chromatographic behaviors which are used to help in identification. In the figure below are plotted the elution profiles (reversed phase) and has and we always recommend and suggest that important finding are validated using standards. Lipids
Lipid are often identified by matching fragmentation data to a library and/or database. Different groups of lipids often behaves similar with respect to elution profile (reversed phase) which can help the identification.