Untargeted Analysis of Polar Metabolites
Un-targeted, or hypothesis generating, analysis of polar metabolites is carried by combining high mass accuracy measurement of precursors with high mass accuracy fragmentation data. Molecules are most often selected based on abundance where after a selected mass is excluded for a chromatographic relevant time period (Data Dependent Acquisition, DDA, or ‘shotgun analysis’). Polar metabolites the analytes are separated using hydrophilic interaction chromatography (HILIC). Currently, the Center using two Data Dependent Acquisition strategies: 1) a pooled sample, composed of each of the samples to be analyzed, is analyzed extensively to create a library of identified molecules. Individual samples are then analyzed in polarity switch mode and pool-identified analytes are mapped to MS1 signals of the samples, or, 2) Each sample is analyzed separately in positive and negative mode and MS/MS data are collected using DDA.
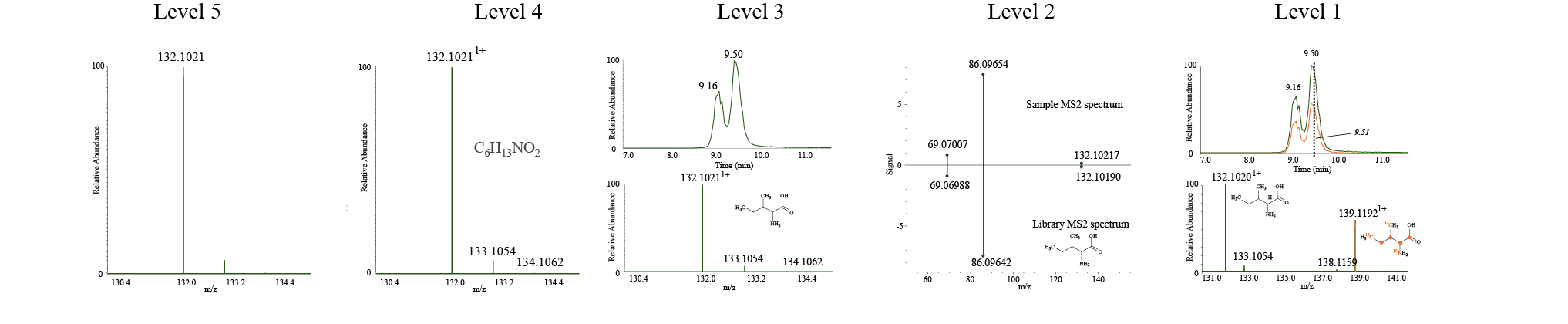
Data are analyzed using Compound Discoverer. Even high mass accuracy/high resolution fragmentation data can be difficult to match and we always recommend and suggest that important finding are validated using standards. The field of metabolomics operates with different levels of identification where Level 1 is the strongest type of identification that is based on standards.
Level 1 through 5 type identification of metabolite identifications levels as described by Schrimpe-Rutledge et al: Level 5: Unique feature, Feature can be compared between samples. Level 4: Features are assigned possible atomic composition based on accurate mono isotopic mass, isotope distribution and charge. Level 3: Tentative structure is assigned based on additional information, for example Retention Time. Level 2: Putative identification based on fragmentation, for example with match to library database. Level 1: Validated identification based on standard which can be heavy isotope labelled.