Quality Control & Amounts
Estimating protein amounts and quality control samples can be challenging. For example, some sample matrices can disturb protein concentration measurements and at times a sample can be contaminated by a very abundant protein that will bias concentration measurements. Left picture below shows a comparison of lystaes for an quantitative protein profiling with two conditions and 5 replicates. Based on the SDS-PAGE analysis, the samples appears comparative. The gel was accompanied by protein conc. measurements.
Furthermore, estimating protein amount using staining intensities can be difficult, especially when based on silver staining(opens in new window) where the intensity is time dependent. We recommend Colloidal Blue(opens in new window) staining (protocol: http://tools.invitrogen.com/content/sfs/manuals/colloidalstain_man.pdf(opens in new window), page 8-9 for Bis-Tris gels) because it delivers maximum staining after 3h which minimizes staining-time bias. Colloidal Blue is less sensitive then silver staining, but more sensitive than Commasie Brilliant Blue(opens in new window). An example of use of SDS-PAGE to QC a proteomics profiling experiment prior to sample drop off is shown below. Importantly, though silver staining is more sensitive when the readout is visualization, our experience is that extraction of peptides from a silver stained in-gel digestion is 5-10 times less efficient when compared to Colloidal Blue(opens in new window).
To assess protein amount, the Proteomics Resource Center suggests to comparing samples to a commercial E.coli lysate. Upon request and availability the PRC will share 20 µg in 20 µL of an E.coli lysate (BioRad) with investigators. The lysate can be helpful in the assessment of protein amount and for general troubleshooting.
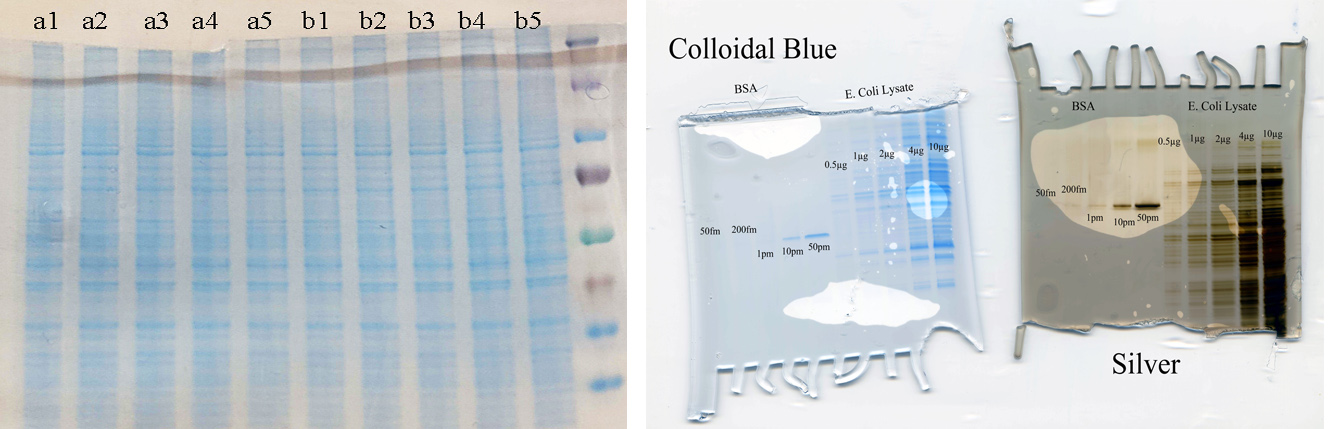
Left: SDS-PAGE used to QC a sample set prior to quantitative proteomics profiling by LC-MS/MS. Right: Comparing staining using a single protein and a full lysate.
Suggested use of E.coli Standard
- Prepare 20 ug E.coli lysate (in 20 uL) such that the total volume, including loading buffer and reducing agents, is 60uL. Follow standard protocol for sample preparation. NOTE: The above volume/concentration is chosen so that it is possible to confidently load 1ug (3uL) in one lane and also be able to load 10 ug (30uL) without overloading the well.
- Load 1 ug of the standard lysate onto one lane and 10 ug onto the adjacent lane.
- Load samples that are to be compared to the E.coli standard onto adjacent lanes.
- Run gel.
For some proteomics experiments a rough estimation of sample amount can be sufficient. However, many applications require assessment of absolute or/and relative sample amounts. Examples are chemical labeling experiments where too much or too little of the labeling reagents can offset the results. Furthermore, during analysis of complex samples overloading of the equipment can lead to sub-optimal chromatography/separation resulting in too few protein identifications.