Laboratory of Molecular Neuro-oncology

Investigator, Howard Hughes Medical Institute

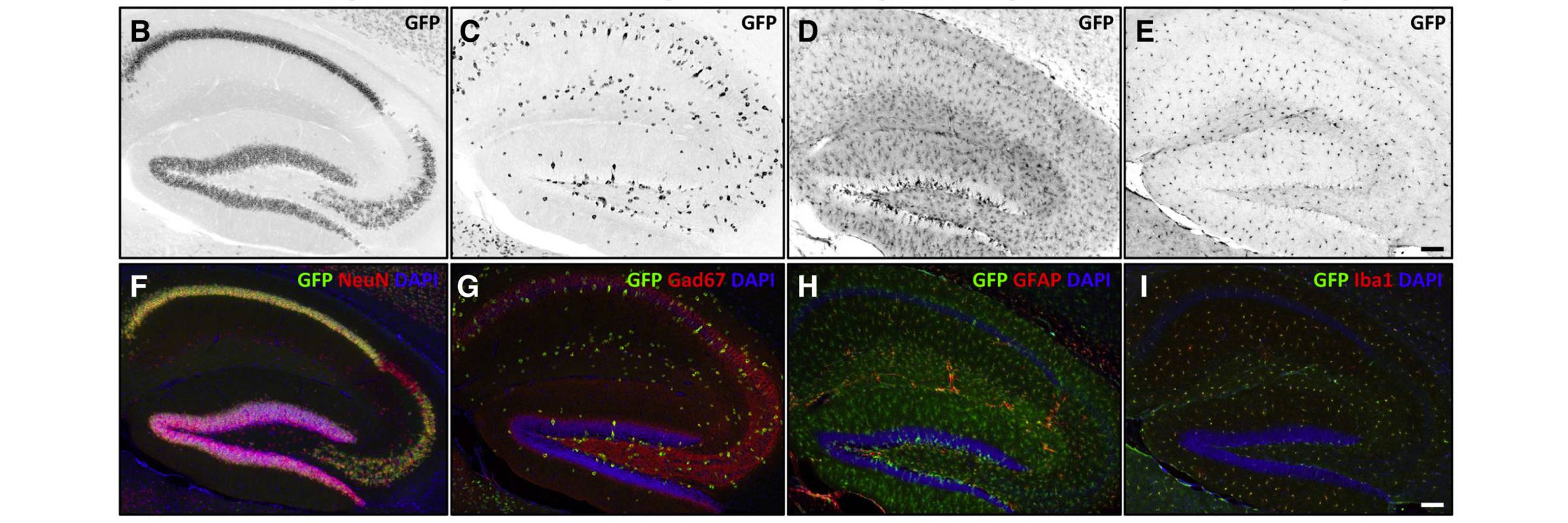
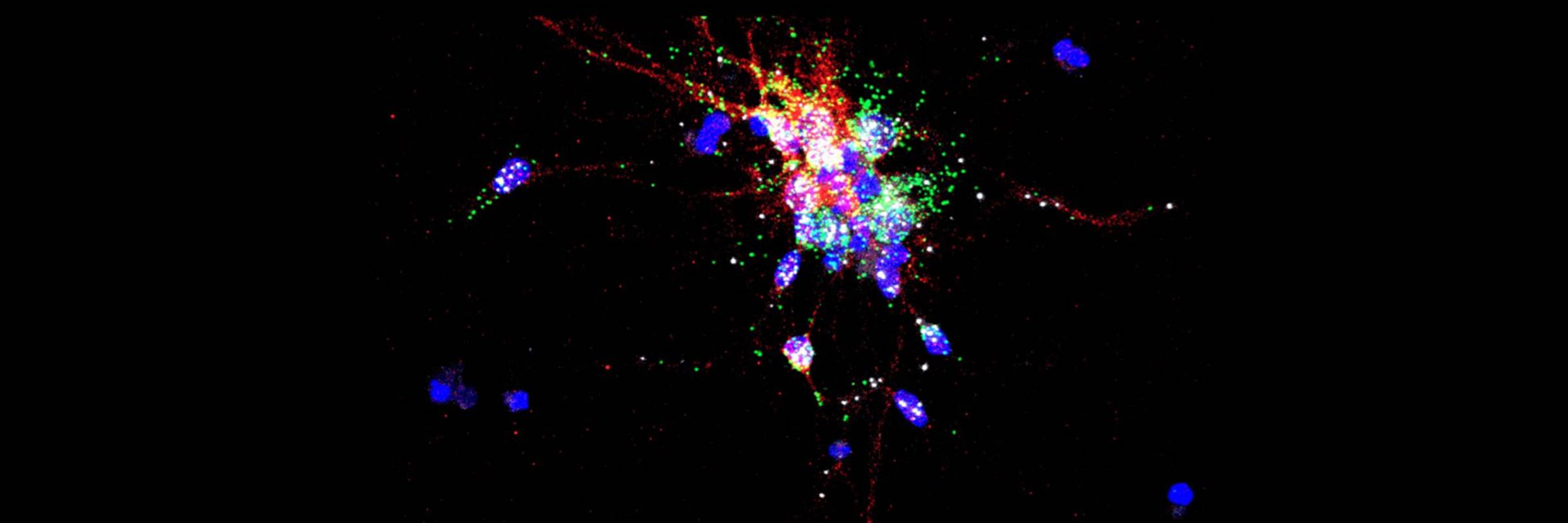
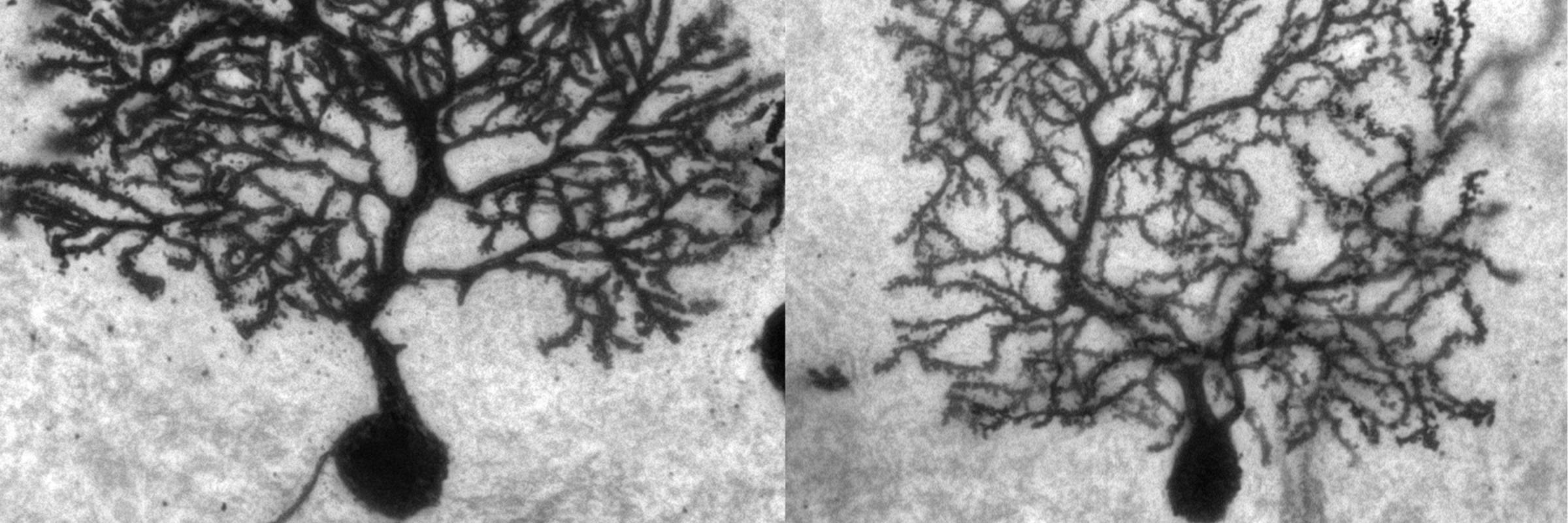

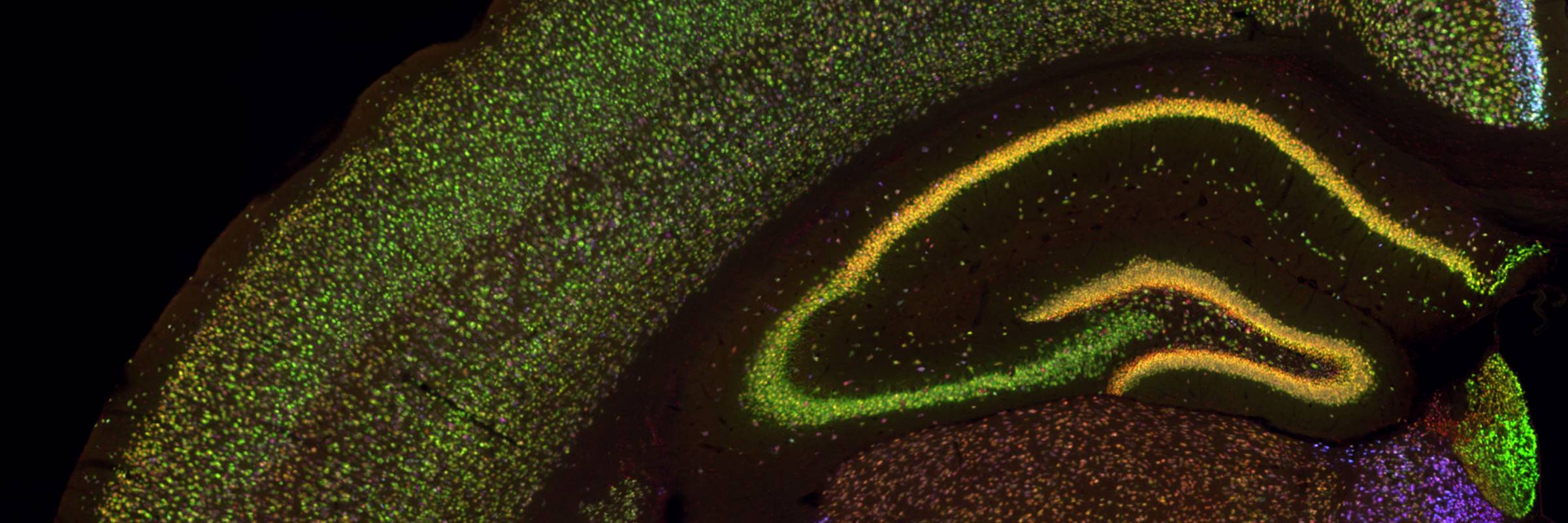
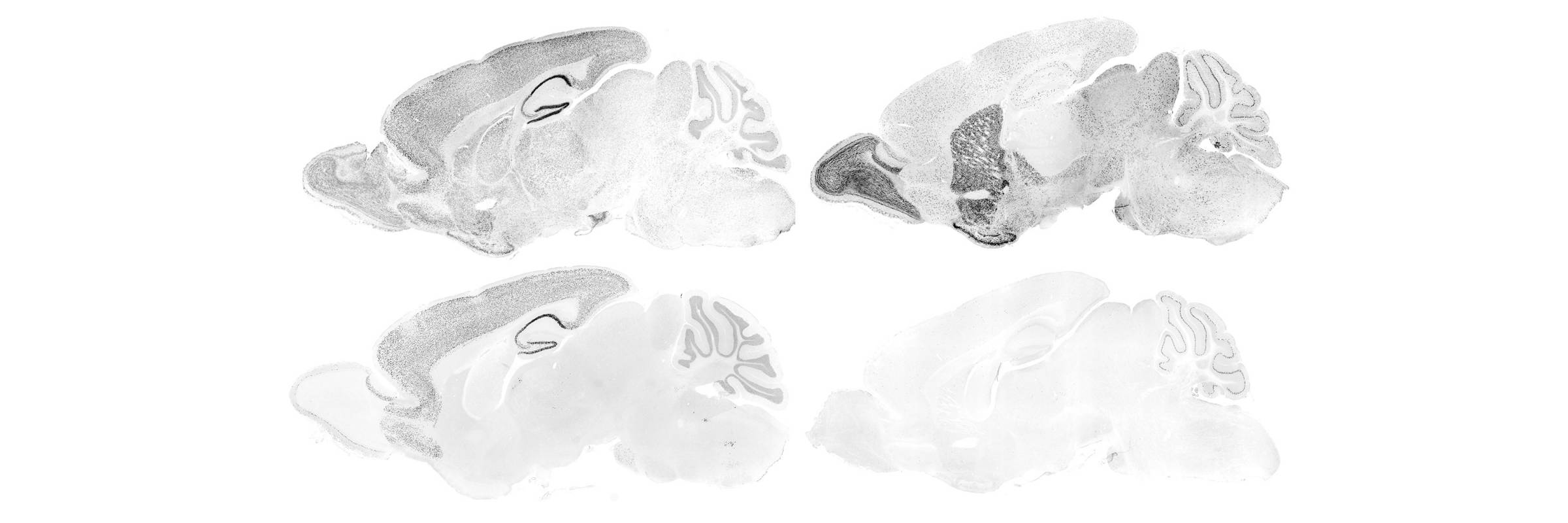
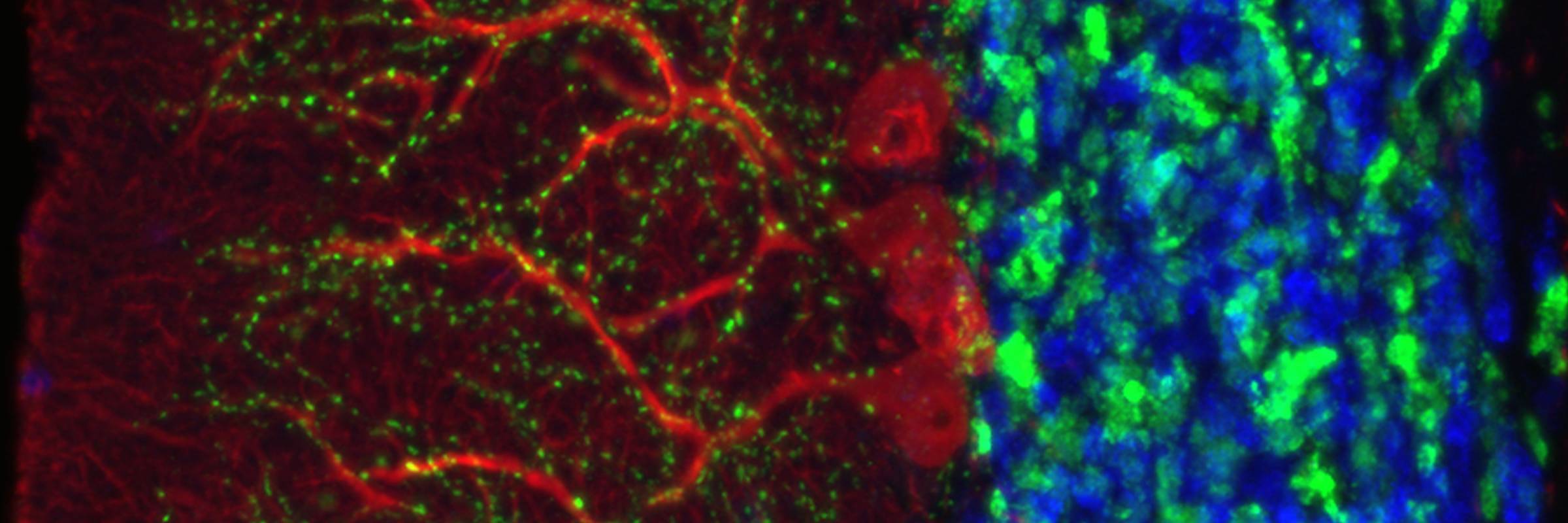
RNA is the driving force of biological complexity, determining which genes any given cell expresses at specific times or under specific circumstances. Darnell developed high-throughput sequencing and crosslinking methods to investigate how RNA binding proteins (RBPs) modulate gene expression. He discovered neuron-specific RNA-binding proteins in the mammalian brain and studies their link to intellectual function and human disease.
Darnell’s interest in RNA and RNA-binding proteins arose from studies of paraneoplastic neurologic disorders (PNDs), a group of rare conditions that affect the nervous system in people with certain tumors—typically breast, ovarian, or lung cancers. These disorders are thought to arise when tumors make proteins normally unique to the brain, triggering an immune response that thwarts the cancer but also causes collateral damage to the nervous system.
Using a combination of biochemical and genetic approaches, Darnell’s lab identified the neuron-specific proteins targeted by the immune system in PNDs. The team discovered that the immune systems of PND patients kill tumor cells with what is essentially an antiviral response: CD8+ killer T cells that recognize the neuron-specific proteins. The lab also found that dying tumor cells further instigate this anti-tumor response and has worked on developing cancer vaccines that mimic this effect. Their work demonstrating that killer T cells mediate naturally occurring human tumor immunity provided key support for the emerging field of immuno-oncology.
Subsequent studies in the Darnell lab explored why tumors induce the expression of neuron-specific proteins, and these proteins’ normal roles in neurons. This work led the lab to discover and explore the function of neuron-specific RNA-binding proteins in the mammalian brain, including Hu (nElavl) and Nova. Additional studies led to the discovery of the function of the related RNA-binding protein FMRP, whose function is lost in fragile X syndrome, the most common inherited form of intellectual disability. The lab further expanded these investigations to include other RNA-binding proteins, such as Argonaute (Ago, which works with microRNAs to regulate messenger RNA), as well as Ptbp2, Mbln2, and Rbfox.
To understand these proteins’ functions in the brain, the lab developed a general and powerful method called cross-linking immunoprecipitation (CLIP), which allowed the team to create precise, genome-wide maps of protein binding sites on RNAs in living tissue. They used CLIP, together with the analysis of knockout mice and bioinformatic approaches, to discover that where a protein binds influences how messenger RNAs are altered through processes called alternative splicing and polyadenylation. Recent computational improvements in the analysis of CLIP maps have allowed single-nucleotide and single cell-type resolution of RNA regulatory sites in different neurons, and robust genome-wide predictions of combinatorial RNA regulation in the non-coding genome.
The lab has revealed how disruptions in RNA-protein interactions contribute to common neurologic disorders, such as that mediated by FMRP in autism spectrum disorders, by Ago in stroke and hepatitis infection, by nElavl in Alzheimer’s disease, and by Nova in axonal guidance and brain development. Studies with Nova and FMRP have led to insight into the balance of neuronal inhibition, excitation, and control of gene expression, leading to new approaches to study neurodegenerative disorders such as Lou Gehrig’s disease and developmental disorders such as epilepsy and autism.